Hijacking strategy mapped for hundreds of viruses

One strategy that viruses use to take over a host cell is to mimic small parts of the cell's proteins called motifs. In a new study coordinated from Uppsala University, researchers have used a new method and doubled the available information on how viruses mimic human binding motifs. The results suggest new targets for the development of antiviral inhibitors. The research is published in the journal Nature Communications.
Viruses are infectious particles that depend on host cells to reproduce. When a virus enters a host cell, it hijacks the cell's processes to produce more virus particles. One type of mechanism used is mimicking the motifs.
Proteins provide structure, transmit signals, and catalyze chemical reactions needed for normal cell function. Proteins themselves are made up of amino acids. The order of the amino acids determines the protein's shape, size, and function. While some regions of a protein fold to form well-defined three-dimensional structures, other regions contain short stretches of amino acids that act as barcodes. These barcodes are called motifs and are recognized and bound by other proteins.
Motif-based protein interactions are crucial for a range of important cellular processes, such as enabling communication between cells, recycling of damaged proteins, and transport of proteins between different cellular compartments. Since motifs are small, it is easy for viral proteins to evolve their own interaction motifs that compete with the cell's protein interactions.
Over the past few decades, several research studies have been conducted on how viruses mimic human binding motifs. Most studies have focused on one virus and one interaction at a time. It has been previously known that interactions are important for how viruses take over our cells, although the information has been limited.
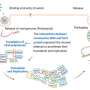
The researchers behind the present study have used a new method to systematically map interactions between human proteins and proteins from hundreds of different viruses, essentially doubling the available information and providing a completely new understanding of viral motif mimicry.
"To date, there has been no research looking at these interactions on a large scale because they are notoriously difficult to identify and study. Our results are the first large-scale attempt to simultaneously identify and characterize motif-dependent interactions from many different virus species," says Filip Mihalič, a researcher at the Department of Medical Biochemistry and Microbiology at Uppsala University and the first author of the article.
The new study also describes how viruses take over the cell's endocytic transport machinery, which is necessary for the uptake of virus particles but also for the transport of the cell's own proteins. Furthermore, the researchers demonstrate that the protein PABP1, which is important for the synthesis of new proteins, is a target for motif-based hijacking. The researchers showed that some viruses use PABP1 and that it is possible to inhibit virus infection by blocking PABP1's function. The results thus suggest that PABP1 could be a possible target for the development of new antiviral drugs.
"We hope that our results will encourage other researchers to pay more attention to this understudied phenomenon. The results we report now are just the tip of the iceberg," says Filip Mihalič.
"In the future, we hope to create a complete map of motif-dependent interactions between viral and human proteins. This would help us understand what happens during virus infection and lead to the identification of new targets for the development of antiviral drugs to combat virus infections and pandemics like COVID-19," says Ylva Ivarsson, a Professor at the Department of Chemistry BMC, Uppsala University.
More information: Filip Mihalič et al, Large-scale phage-based screening reveals extensive pan-viral mimicry of host short linear motifs, Nature Communications (2023).
Journal information: Nature Communications
Provided by Swedish Research Council