How two key proteins maintain optimal pH within the Golgi apparatus

Lisa Lock
scientific editor

Robert Egan
associate editor

Our cells carry out hundreds of thousands of important functions every day, all of which are carefully orchestrated at a microscopic scale. However, the precise mechanisms that influence the luminal pH鈥攐ne key component that allows cells to function properly鈥攐f each organelle are unclear.
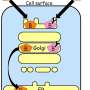
In a study last month in the Proceedings of the National Academy of Sciences, researchers from The University of Osaka have revealed that two closely related proteins help maintain proper conditions within key cellular organelles. These key proteins regulate the environment of the Golgi and trans-Golgi network, which are responsible for modifying proteins to make sure they are functional.
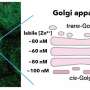
Eukaryotic cells contain a wide variety of organelles, which are sub-compartments in the cells that perform specialized functions that require specific conditions. The luminal pH within these sub-compartments is crucial in enabling them to carry out essential processes like modifying newly created proteins, such as glycosylation.
"Some proteins are known to be associated with proton pumps that regulate the pH within specific organelles," says lead author, Shin-ichiro Yoshimura. "However, the precise mechanisms by which these proteins determine the interior pH of each organelle are not fully understood."
To address this, the researchers searched for proteins that interact with Rab6, a member of the Rab family of proteins. This group of proteins is known to regulate membrane trafficking and define the identity of organelles through major activities, such as vesicular transport.
"The results were very clear," explains Akihiro Harada, senior author. "We found that the Rab-binding proteins Oxr1 and Ncoa7 were specifically located at the Golgi and trans-Golgi network membranes, where they prevented excessive acidification by inhibiting the activity of the vacuolar-type proton pump."
V-ATPase works inside eukaryotic cells to acidify their components, and transport protons across plasma membranes. When Oxr1 and Ncoa7 function was inhibited, V-ATPase was able to significantly increase the interior acidity of the interior of the Golgi and trans-Golgi network.
In addition, the cells showed defects in protein glycosylation, a major enzymatic reaction performed in the Golgi and trans-Golgi network. Defects in glycosylation poses many challenges to cell function as the process ensures cells can maintain their important physiological functions.
"Taken together, our findings suggest that Oxr1 and Ncoa7 regulate V-ATPase at the Golgi apparatus and trans-Golgi network to maintain optimal luminal pH for enzymatic activity," says Yoshimura.
Given that congenital disorders of glycosylation are caused by mutations in genes involved in glycoprotein and lipid synthesis, such as V-ATPase, these findings provide new insight into precisely how these diseases occur.
They may also explain why previous studies in humans with OXR1 deficiency and mice lacking Oxr1 and Ncoa7 have shown abnormal lysosomal function, which can lead to a build-up of undigested materials and lead to cellular dysfunction.
More information: Shin-ichiro Yoshimura et al, Oxr1 and Ncoa7 regulate V-ATPase to achieve optimal pH for glycosylation within the Golgi apparatus and trans-Golgi network, Proceedings of the National Academy of Sciences (2025).
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Osaka