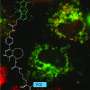
Nanobody-guided approach enables efficient fluorescent labeling of endogenous proteins

Lisa Lock
scientific editor

Robert Egan
associate editor

A research team led by Prof. Xu Pingyong from the Institute of Biophysics of the Chinese Academy of Sciences has developed an innovative approach to visualize and rapidly screen small peptide knockins. The new approach, termed ALFA Nanobody-guided Endogenous Labeling (ANGEL), solves a long-standing problem of high-throughput screening for nonfluorescent small peptide knockins. Results were in Nature Chemical Biology on August 29.
Due to their small size (~15 kDa), high stability, and strong binding affinity, nanobodies have emerged as powerful tools in molecular biology. When fused with fluorescent proteins to form chromobodies, nanobodies enable multicolor live-cell labeling, super-resolution imaging, and real-time studies of protein dynamics. However, conventional chromobody development is laborious and time-consuming.
According to the researchers, ANGEL is built upon the ALFA tag, a rationally designed 13-amino acid peptide that forms a stable 伪-helix. The ALFA tag is smaller than most conventional linear epitope tags and exhibits excellent chemical stability.
They discovered that the stability of NbALFA, a high-affinity nanobody specific to the ALFA tag, depends on the presence of the tag. In the absence of ALFA, NbALFA undergoes partial degradation. However, increasing intracellular ALFA levels stabilize NbALFA and enhance its fluorescence signal.
To harness this property, the researchers constructed stable cell lines expressing NbALFA fused to a fluorescent protein and integrated it into the genome to ensure uniform expression in the absence of the ALFA tag.
Using CRISPR-mediated gene editing, they then precisely inserted the ALFA tag into target loci and monitored changes in NbALFA fluorescence to efficiently identify successfully edited cells.
The ANGEL technique demonstrated remarkable versatility, successfully labeling a wide range of endogenous proteins鈥攊ncluding CKAP4, SEC61B, RTN4, Vimentin, nucleoporins NUP96 and NUP35, histone H2BC21, CBX1, Lamin A/C, Actin, and even the nuclear speckle core protein SON with a molecular weight of 264 kDa.
Through fluorescent nanobody-based multicolor labeling, ANGEL can image across various tissue depths and is compatible with multiple microscopy modalities. More importantly, ANGEL provides a real-time, reliable, and streamlined platform for studying the biological functions of endogenous proteins under native regulatory conditions, overcoming the limitations of traditional overexpression systems.
This work opens new avenues for protein function research, dynamic cellular imaging, and drug discovery, establishing ANGEL as a next-generation platform for precision protein labeling and functional analysis.
More information: Zhe Wang et al, ALFA nanobody-guided endogenous labeling, Nature Chemical Biology (2025).
Journal information: Nature Chemical Biology
Provided by Chinese Academy of Sciences